

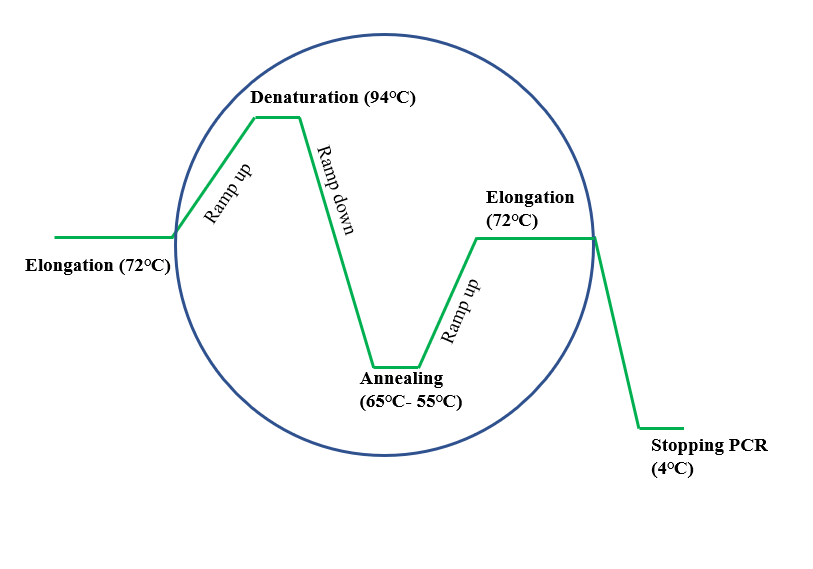
Comparison of two methods for purifying PCR products prior to sequencing showed that purification using MultiScreen 384-PCR filter plates had an advantage over ethanol purification because greater numbers of sequencing reactions could be performed from comparable volumes of PCR reactions. The hemi-nested TD PCR increased both specificity and yield by high and low annealing temperatures in two consecutive amplifications. For the hemi-nested TD PCR, primers were designed based on the known sequences obtained and/or published. Two or more primer-template mismatches reduced PCR product yield approximately from 2-fold to 10-fold compared to PCR product yield without the primer-template mismatch. These mismatches included the transition mispairs G:T, T:G, A:C and the transversion mispairs A:A, A:G, G:G, G:A. Single or multiple mismatches were observed at 5' terminal, internal and the penultimate position, respectively. The effects of the universal primer-template mismatches on the efficiency of standard PCR amplification were investigated after assembly of sequences from different primers amplifying the same BAC clones. Resultsįor the primer-template mismatch PCR, the universal primers were designed based on conserved gene coding regions of consensus sequences. Here we report on an efficient approach for rapid isolation and sequencing of the low molecular weight glutenin subunit gene family from the 'Glenlea' wheat BAC library via primer-template mismatch PCR using universal primers, primer walking using hemi-nested touchdown (TD) PCR, and followed by direct sequencing of PCR products.
Bacterial artificial chromosome (BAC) libraries can be useful for such work. Isolation of a large number of gene sequences from complex gene families with a high level of gene sequence identity from genomic DNA is therefore difficult and time-consuming. Hexaploid wheat ( Triticum aestivum L.) possesses a large genome that contains 1.6 × 10 10 bp of DNA.


 0 kommentar(er)
0 kommentar(er)
